-
- Downloads
Added demo!
Showing
- .gitignore 2 additions, 0 deletions.gitignore
- demos/Pyramid Notebook Example.ipynb 1099 additions, 0 deletionsdemos/Pyramid Notebook Example.ipynb
- demos/files/magdata.hdf5 0 additions, 0 deletionsdemos/files/magdata.hdf5
- demos/files/phase.unf 0 additions, 0 deletionsdemos/files/phase.unf
- demos/files/phasemap.hdf5 0 additions, 0 deletionsdemos/files/phasemap.hdf5
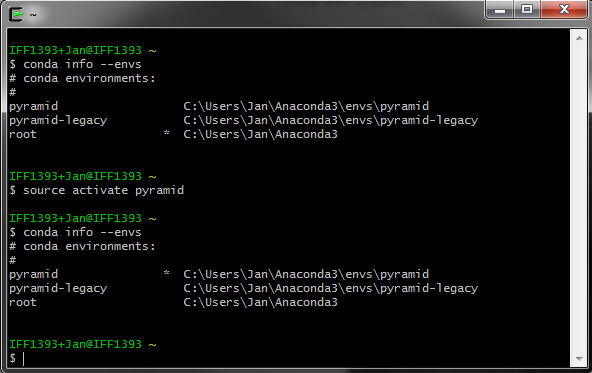
- demos/images/activate.png 0 additions, 0 deletionsdemos/images/activate.png
- demos/images/addsource.png 0 additions, 0 deletionsdemos/images/addsource.png
- demos/images/confidence.png 0 additions, 0 deletionsdemos/images/confidence.png
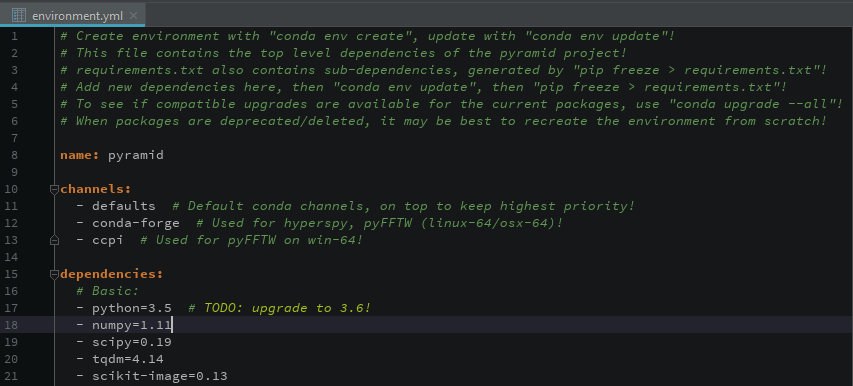
- demos/images/environment.png 0 additions, 0 deletionsdemos/images/environment.png
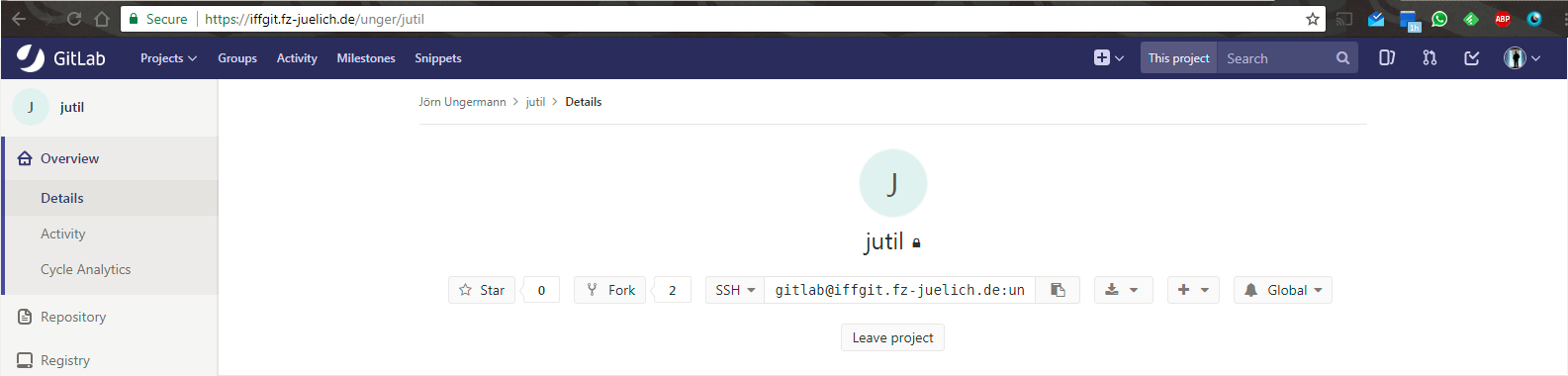
- demos/images/jutil-gitlab.png 0 additions, 0 deletionsdemos/images/jutil-gitlab.png
- demos/images/mask.png 0 additions, 0 deletionsdemos/images/mask.png
- demos/images/phasemap-attributes.png 0 additions, 0 deletionsdemos/images/phasemap-attributes.png
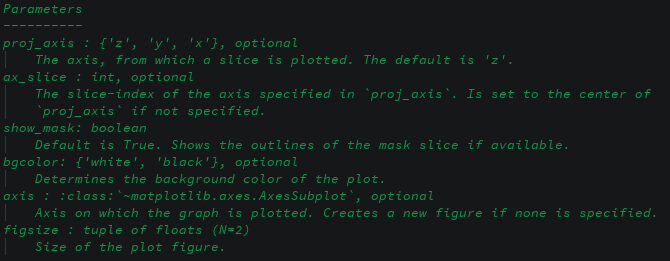
- demos/images/plot-field-params.png 0 additions, 0 deletionsdemos/images/plot-field-params.png
- demos/images/plot-holo-params.png 0 additions, 0 deletionsdemos/images/plot-holo-params.png
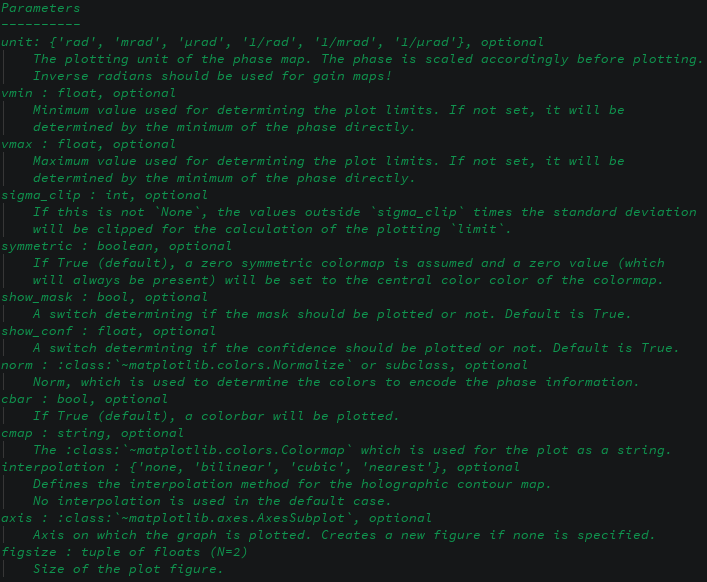
- demos/images/plot-phase-params.png 0 additions, 0 deletionsdemos/images/plot-phase-params.png
- demos/images/plot-quiver-params.png 0 additions, 0 deletionsdemos/images/plot-quiver-params.png
- demos/images/pm-params.png 0 additions, 0 deletionsdemos/images/pm-params.png
- demos/images/pyramid-gitlab.png 0 additions, 0 deletionsdemos/images/pyramid-gitlab.png
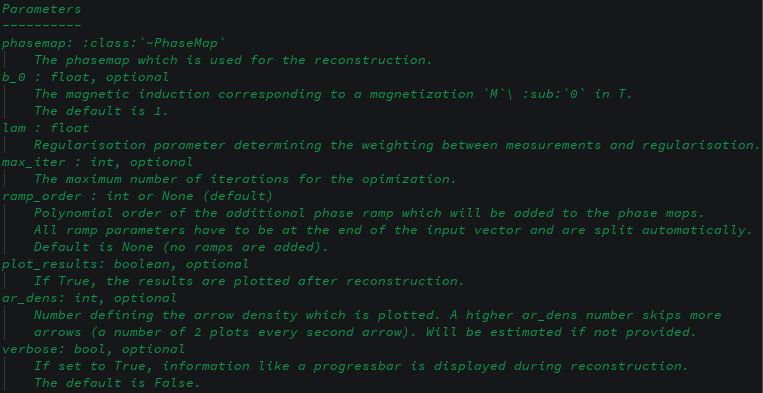
- demos/images/rec2d-params.png 0 additions, 0 deletionsdemos/images/rec2d-params.png
- demos/images/vectordata-attributes.png 0 additions, 0 deletionsdemos/images/vectordata-attributes.png
demos/Pyramid Notebook Example.ipynb
0 → 100644
demos/files/magdata.hdf5
0 → 100644
File added
demos/files/phase.unf
0 → 100644
File added
demos/files/phasemap.hdf5
0 → 100644
File added
demos/images/activate.png
0 → 100644
32.4 KiB
demos/images/addsource.png
0 → 100644
56 KiB
demos/images/confidence.png
0 → 100644
1.56 KiB
demos/images/environment.png
0 → 100644
57.1 KiB
demos/images/jutil-gitlab.png
0 → 100644
63.1 KiB
demos/images/mask.png
0 → 100644
2.37 KiB
demos/images/phasemap-attributes.png
0 → 100644
34.6 KiB
demos/images/plot-field-params.png
0 → 100644
34.8 KiB
demos/images/plot-holo-params.png
0 → 100644
34.2 KiB
demos/images/plot-phase-params.png
0 → 100644
88.8 KiB
demos/images/plot-quiver-params.png
0 → 100644
96.5 KiB
demos/images/pm-params.png
0 → 100644
31.3 KiB
demos/images/pyramid-gitlab.png
0 → 100644
63.1 KiB
demos/images/rec2d-params.png
0 → 100644
54.7 KiB
demos/images/vectordata-attributes.png
0 → 100644
14.4 KiB